BLOSUM
BLOSUM ( blocks substitution matrix ) is an evidence-based substitution matrix, which is used for sequence alignment of proteins and in addition to the Point Accepted Mutation matrix ( PAM matrix ) plays an important role in bioinformatics. The BLOSUM was developed in 1992 by Jorja G. Henikoff and Henikoff Steven. There are different templates for different evolutionary distances.
Calculation
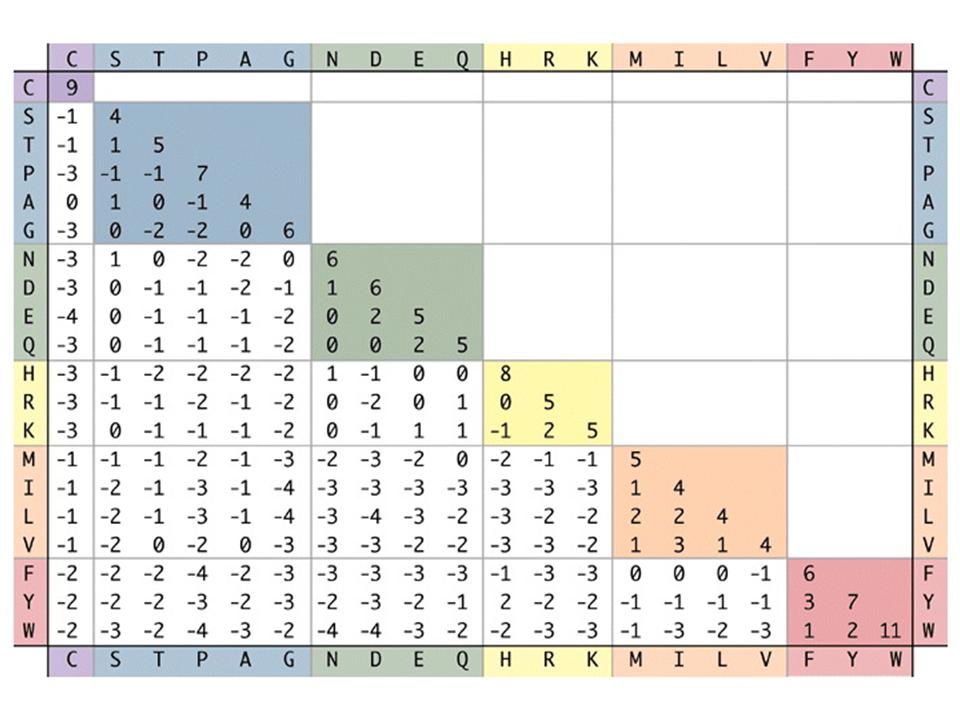
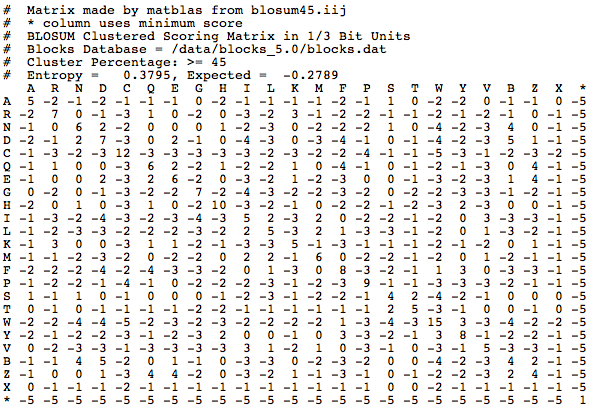
BLOSUM uses individual blocks ( without gaps) within the sequences of homologous proteins that are compared. There are various BLOSUM which are designed for different areas of application. So BLOSUM are suitable with high numbers such as BLOSUM80 for evolutionary closely related proteins and those with low numbers like blosum45 suitable for highly divergent proteins. The authors of the BLOSUM matrix have according to the number all the blocks that had a higher sequence identity than the fixed percentage, combined into a single sequence ( clustering ) in order to reduce the influence of closely related blocks. As BLOSUM80 for all sequences having more than 80% sequence identity have been joined together, so that all of the remaining sequences were compared to each other less than 80% identity. Be entered in the matrix, the log -odds values:
The probability i and j to find the amino acids in an alignment and or is the frequency of amino acids at all. is a normalization factor, the values are rounded to whole numbers. Thus, the logarithm is greater than zero and a positive score results when the two amino acids are commonly found in an alignment than would be expected by chance. Thus, for example, The value for a substitution of tryptophan to tyrosine in the BLOSUM62 2 greater than zero, which means that tryptophan to tyrosine ( and vice versa) mutated frequently than would be expected by chance - This also due to the similar physical and chemical characteristics of the two amino acids sense. The largest score is usually observed, however, for the identity, so has a tryptophan, which is a tryptophan, a score of 11, and a tyrosine, which is a tyrosine, a score of 7 The advantage of log odds is that these can be added instead of multiplying as normally probabilities and this makes the computation easier numerically. The probability itself can be easily recovered by the score will exponenziert.
Use
BLOSUM high speeds ( for example, BLOSUM80 ) are used for the comparison of closely related sequences, as BLOSUM is used with low numbers for the comparison of distantly related proteins. Often an alignment of two sequences using the BLOSUM is evaluated. So, for example, has the following alignment
EKNGFPA | | | EMQGRWA with the BLOSUM62 the score 7 The algorithms that are either global ( Needleman & Wunsch ) or local (Smith & Waterman ) pairwise perform sequence alignment using, for protein sequences often than BLOSUM substitution matrix, but this can be chosen freely. The algorithms BLAST or FASTA, which search a database for a particular sequence use, often BLOSUM for protein searches. The user is often not interested in exact matches and albeit related, but not identical proteins are looking for, then it can be evaluated using the BLOSUM whether the alignment to a particular protein in the database is significant or not.